All-department team publishes paper in Computational and Structural Biotechnology Journal
Congratulations to former visiting student Chia-Jung "Charlene" Chang (PhD candidate in biomedical engineering at National Cheng Kung University), research assistant professor Chih-Yuan Hsu, and professors Qi Liu and Yu Shyr on the publication of "VICTOR: Validation and inspection of cell type annotation through optimal regression." The article on this new method appeared online ahead of print on October 15 and will go to press as part of Computational and Structural Biotechnology Journal's December issue. As described in the paper's abstract:
Single-cell RNA sequencing provides unprecedent opportunities to explore the heterogeneity and dynamics inherent in cellular biology. An essential step in the data analysis involves the automatic annotation of cells. Despite development of numerous tools for automated cell annotation, assessing the reliability of predicted annotations remains challenging, particularly for rare and unknown cell types. Here, we introduce VICTOR: Validation and inspection of cell type annotation through optimal regression. VICTOR aims to gauge the confidence of cell annotations by an elastic-net regularized regression with optimal thresholds. We demonstrated that VICTOR performed well in identifying inaccurate annotations, surpassing existing methods in diagnostic ability across various single-cell datasets, including within-platform, cross-platform, cross-studies, and cross-omics settings.
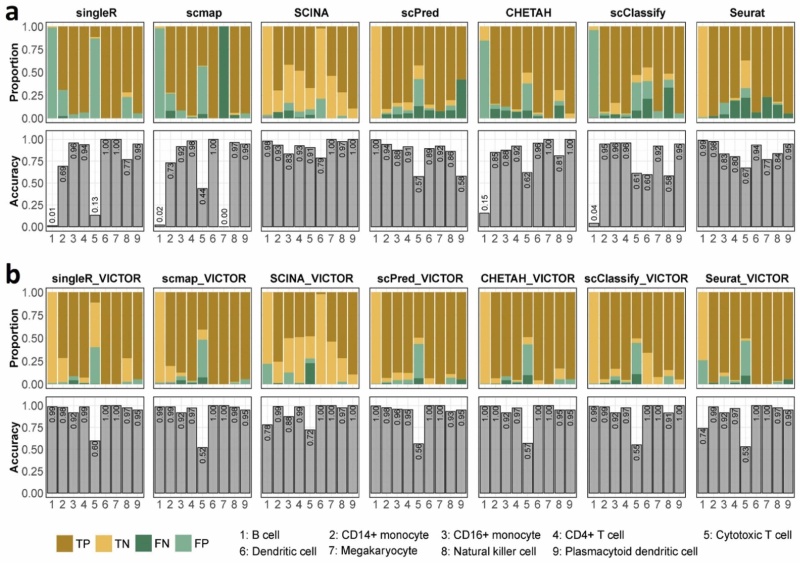
Figure 1 in the paper provides an " One example in diagnosing the reliability of cell annotations. a) diagnostic performance of singleR, scmap, SCINA, scPred, CHETAH, scClassify, and Seurat. b) diagnostic performance of VICTOR when applied to annotations from singleR, scmap, SCINA, scPred, CHETAH, scClassify, and Seurat.

Chih-Yuan Hsu is first author of all-department paper in Bioinformatics
Congratulations to research assistant professor Chih-Yuan Hsu (first author) and professors Qi Liu and Yu Shyr (corresponding authors) on the publication of A distribution-free and analytic method for power and sample size calculation in single-cell differential expression in Bioinformatics. The paper addresses potential deviations from true data distribution in power and sample size calculation by offering a new method, scPS, that "stands out by making no assumptions about the data distribution and considering cell-cell correlations within individual samples. scPS is a rapid and powerful approach for designing experiments in single-cell differential expression analysis." There are web versions of scPS's independent two-group comparison and paired-group comparison tools as well.

An illustration of sample sizes and cells per sample in aiming toward a power of .80. This is Figure 1.2-1 in the independent file set for the scPS package on GitHub.
First-authored paper in Genome Biology by Jia Li
Congratulations to postdoctoral fellow Jia Li on the publication of "aKNNO: single-cell and spatial transcriptomics clustering with an optimized adaptive k-nearest neighbor graph" in Genome Biology on August 1, with professors Yu Shyr and Qi Liu as corresponding authors. The paper offers a new method "to simultaneously identify abundant and rare cell types based on an adaptive k-nearest neighbor graph with optimization," doing so "more accurately than general and specialized methods." {aKKNO} and related tutorials are free and available on GitHub and Zenodo.
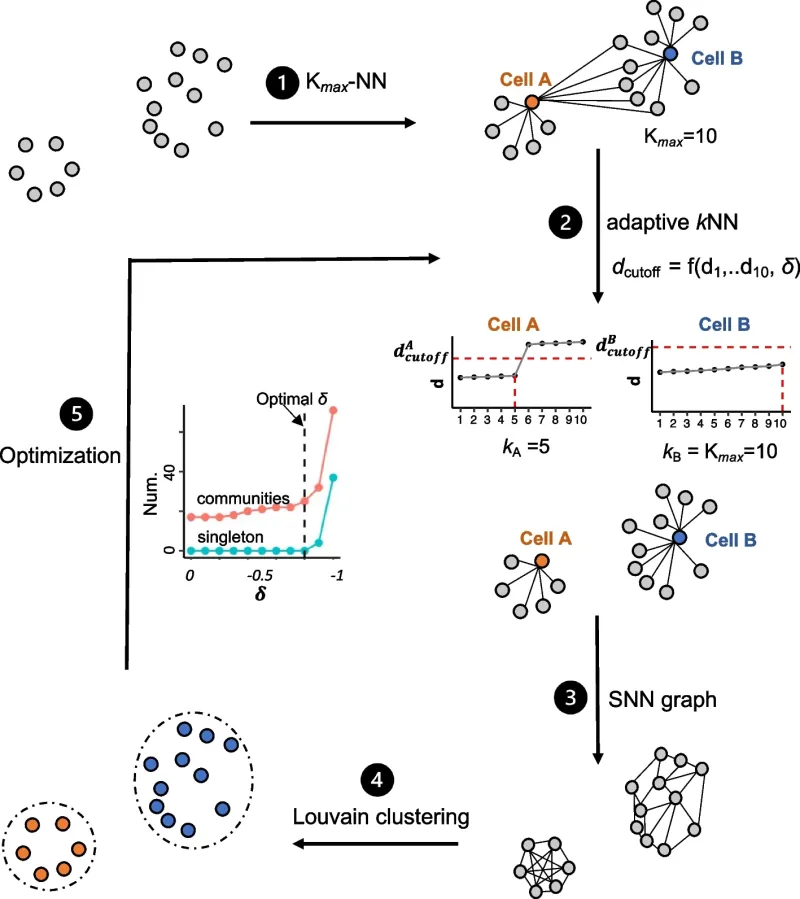
Figure 1 in the paper provides an overview of how aKNNO works.