Bill Dupont is first author of JCO Precision Oncology paper
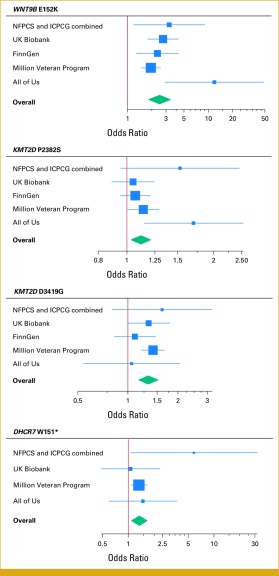
Congratulations to professor of biostatistics and health policy William D. Dupont, PhD, FASA, on the publication of Coding Variants of the Genitourinary Development Gene WNT9B Carry High Risk for Prostate Cancer, published January 28 in JCO Precision Oncology in collaboration with Angela Jones (VANTAGE), Jeffrey R. Smith (Department of Medicine), and the VA Million Veteran Program. Dr. Dupont and Dr. Smith are also affiliated with the Medical Research Service of the Tennessee Valley Healthcare System, and the study was supported by grants from the Veterans Health Administration, the V Foundation for Cancer Research, and the National Institutes of Health, with data drawn from the NIH's All of Us program, the Nashville Familial Prostate Cancer Study (NFPCS), the International Consortium for Prostate Cancer Genetics (ICPCG), UK Biobank, and FinnGen.
The goal of the study was to detect and confirm potential links between specific genes and hereditary prostate cancer. Dr. Dupont and his colleagues used genome-wide single-allele and identity-by-descent analytic approaches to identify pathogenic variants (i.e., mutations) that prostate cancer patients might have in common, and then attempted to replicate their observations with data from the biobanks listed above, confirming the association of two genes — HOXB13 and WNT9B — with high risk of prostate cancer. The paper is the first to discuss WNT9B as a factor in the transmission of prostate cancer. Both genes are vital to prostate development, which suggests that other genes with that characteristic may likewise elevate an individual's risk of developing genitourinary cancer. This new knowledge can help biomedical researchers develop ways to prevent and treat this type of cancer.
Study in multiancestry populations uncovers new insights into breast cancer genetics
Associate professor Ran Tao co-authored this study.
Jing Wang wins 2024 Faculty Development Award
Congratulations to research assistant professor Jing Wang, the 2024 winner of our department's Faculty Development Award. Her proposal was deemed the best in a field of highly competitive submissions for a one-year pilot grant. Dr. Wang will use this funding to support an investigation that she hopes to build into a larger project: “Taking advantage of the dramatically increasing number of single-cell transcriptomics and epigenomics studies, the project aims to develop cell-type-specific enhancer-mediated regulatory maps for functionally interpreting and prioritizing enhancer risk variants and causal genes.” Dr. Wang is shown here describing her project at the August 2024 faculty meeting, when the award was announced. Her accomplishments include first-authorship of a 2023 paper in Cancers (Basel) on small RNA profiling in human extracellular vesicles,
Breast cancer risk variants identified for women of African ancestry
This Nature Genetics paper was co-authored by associate professor Ran Tao.